Of your input list, 11 genes are known or unknown but not ambiguous. Identifiers unknown to the annotation provider are still included in the statistics. The total number of genes used to calculate the background distribution of GO terms is 7166. Displaying 17 terms out of a total of 25 found.
Result Table| Terms from the Process Ontology of gene_association.sgd with p-value <= 0.01 | | Gene Ontology term | Cluster frequency | Genome frequency | Corrected
P-value | FDR | False
Positives | Genes annotated to the term |
|---|
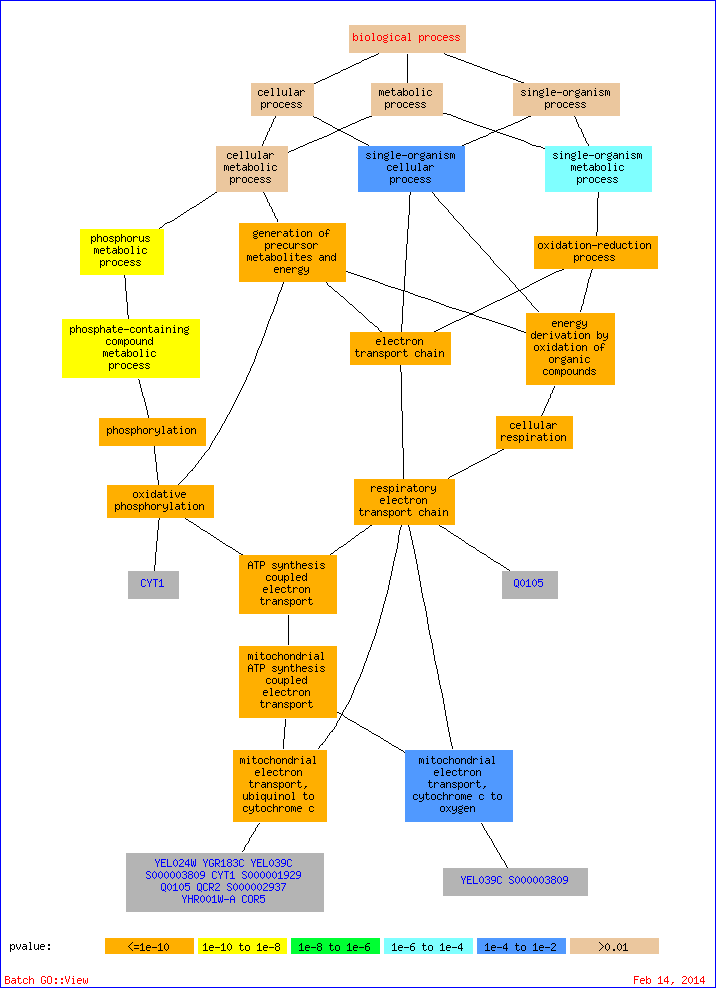
| mitochondrial electron transport, ubiquinol to cytochrome c | 11 of 11 genes, 100.0% | 11 of 7166 genes, 0.2% | 3.92e-34 | 0.00% | 0.00 | YEL024W, YGR183C, YEL039C, S000003809, CYT1, S000001929, Q0105, QCR2, S000002937, YHR001W-A, COR5 | | ATP synthesis coupled electron transport | 11 of 11 genes, 100.0% | 28 of 7166 genes, 0.4% | 8.43e-27 | 0.00% | 0.00 | YEL024W, YGR183C, YEL039C, S000003809, CYT1, S000001929, Q0105, QCR2, S000002937, YHR001W-A, COR5 | | mitochondrial ATP synthesis coupled electron transport | 11 of 11 genes, 100.0% | 28 of 7166 genes, 0.4% | 8.43e-27 | 0.00% | 0.00 | YEL024W, YGR183C, YEL039C, S000003809, CYT1, S000001929, Q0105, QCR2, S000002937, YHR001W-A, COR5 | | oxidative phosphorylation | 11 of 11 genes, 100.0% | 29 of 7166 genes, 0.4% | 1.35e-26 | 0.00% | 0.00 | YEL024W, YGR183C, YEL039C, S000003809, CYT1, S000001929, Q0105, QCR2, S000002937, YHR001W-A, COR5 | | respiratory electron transport chain | 11 of 11 genes, 100.0% | 30 of 7166 genes, 0.4% | 2.14e-26 | 0.00% | 0.00 | YEL024W, YGR183C, YEL039C, S000003809, CYT1, S000001929, Q0105, QCR2, S000002937, YHR001W-A, COR5 | | electron transport chain | 11 of 11 genes, 100.0% | 31 of 7166 genes, 0.4% | 3.32e-26 | 0.00% | 0.00 | YEL024W, YGR183C, YEL039C, S000003809, CYT1, S000001929, Q0105, QCR2, S000002937, YHR001W-A, COR5 | | cellular respiration | 11 of 11 genes, 100.0% | 109 of 7166 genes, 1.5% | 1.50e-19 | 0.00% | 0.00 | YEL024W, YGR183C, YEL039C, S000003809, CYT1, S000001929, Q0105, QCR2, S000002937, YHR001W-A, COR5 | | energy derivation by oxidation of organic compounds | 11 of 11 genes, 100.0% | 157 of 7166 genes, 2.2% | 9.82e-18 | 0.00% | 0.00 | YEL024W, YGR183C, YEL039C, S000003809, CYT1, S000001929, Q0105, QCR2, S000002937, YHR001W-A, COR5 | | generation of precursor metabolites and energy | 11 of 11 genes, 100.0% | 190 of 7166 genes, 2.7% | 8.53e-17 | 0.00% | 0.00 | YEL024W, YGR183C, YEL039C, S000003809, CYT1, S000001929, Q0105, QCR2, S000002937, YHR001W-A, COR5 | | phosphorylation | 11 of 11 genes, 100.0% | 325 of 7166 genes, 4.5% | 3.54e-14 | 0.00% | 0.00 | YEL024W, YGR183C, YEL039C, S000003809, CYT1, S000001929, Q0105, QCR2, S000002937, YHR001W-A, COR5 | | aerobic respiration | 8 of 11 genes, 72.7% | 85 of 7166 genes, 1.2% | 1.12e-12 | 0.00% | 0.00 | Q0105, YEL024W, S000002937, QCR2, YGR183C, YHR001W-A, COR5, S000001929 | | oxidation-reduction process | 11 of 11 genes, 100.0% | 447 of 7166 genes, 6.2% | 1.23e-12 | 0.00% | 0.00 | YEL024W, YGR183C, YEL039C, S000003809, CYT1, S000001929, Q0105, QCR2, S000002937, YHR001W-A, COR5 | | phosphate-containing compound metabolic process | 11 of 11 genes, 100.0% | 816 of 7166 genes, 11.4% | 9.82e-10 | 0.00% | 0.00 | YEL024W, YGR183C, YEL039C, S000003809, CYT1, S000001929, Q0105, QCR2, S000002937, YHR001W-A, COR5 | | phosphorus metabolic process | 11 of 11 genes, 100.0% | 841 of 7166 genes, 11.7% | 1.37e-09 | 0.00% | 0.00 | YEL024W, YGR183C, YEL039C, S000003809, CYT1, S000001929, Q0105, QCR2, S000002937, YHR001W-A, COR5 | | single-organism metabolic process | 11 of 11 genes, 100.0% | 2139 of 7166 genes, 29.8% | 4.11e-05 | 0.00% | 0.00 | YEL024W, YGR183C, YEL039C, S000003809, CYT1, S000001929, Q0105, QCR2, S000002937, YHR001W-A, COR5 | | mitochondrial electron transport, cytochrome c to oxygen | 2 of 11 genes, 18.2% | 12 of 7166 genes, 0.2% | 0.00350 | 0.75% | 0.12 | YEL039C, S000003809 | | single-organism cellular process | 11 of 11 genes, 100.0% | 3280 of 7166 genes, 45.8% | 0.00457 | 0.71% | 0.12 | YEL024W, YGR183C, YEL039C, S000003809, CYT1, S000001929, Q0105, QCR2, S000002937, YHR001W-A, COR5 |
Of your input list, 11 genes are known or unknown but not ambiguous. Identifiers unknown to the annotation provider are still included in the statistics. The total number of genes used to calculate the background distribution of GO terms is 7166. Displaying 17 terms out of a total of 25 found.
Nodes in the graph are color-coded according to their p-value. Genes in the GO tree are associated with the GO term(s) to which they are directly annotated. To control the graph size, only significant hits with a p-value <= 0.01 and terms descended from them are included.
|
|